|
I am an Assistant Professor with the School of Software, Northwestern Polytechnical University since July 2020. I received my Ph.D degree and Bachelor's degree from the Department of Computer Science, City University of Hong Kong. |

|
|
I'm interested in bioinformatics, single-cell sequencing, machine learning, data mining, algorithm design, software development. |
|
|
|
|
|
|
|
|
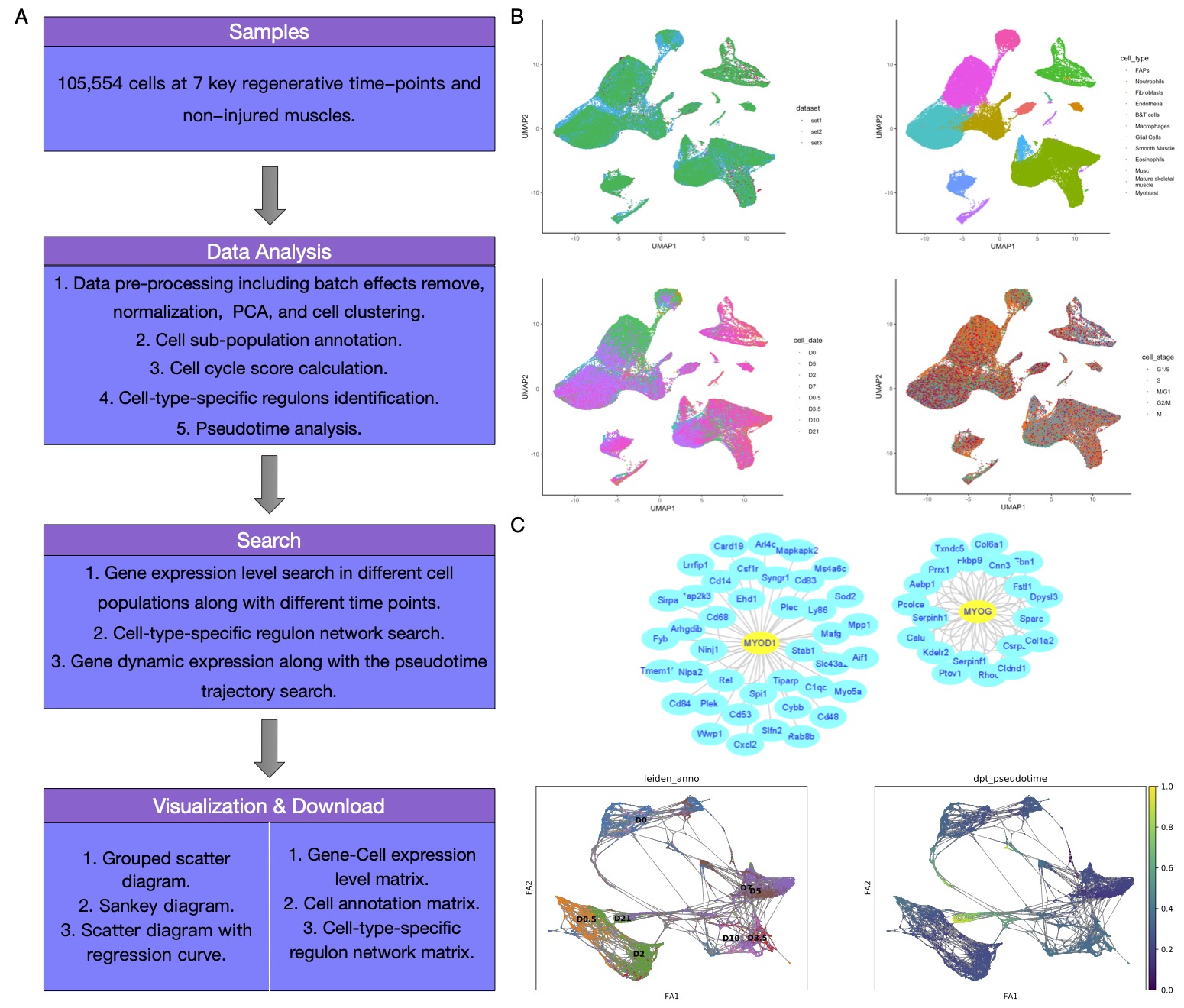
|
|
|
|
|
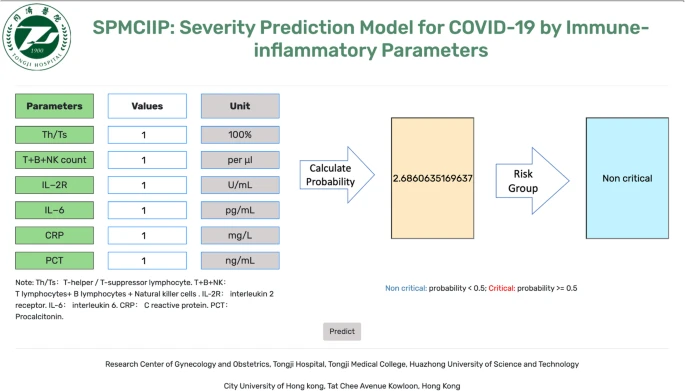
|
|
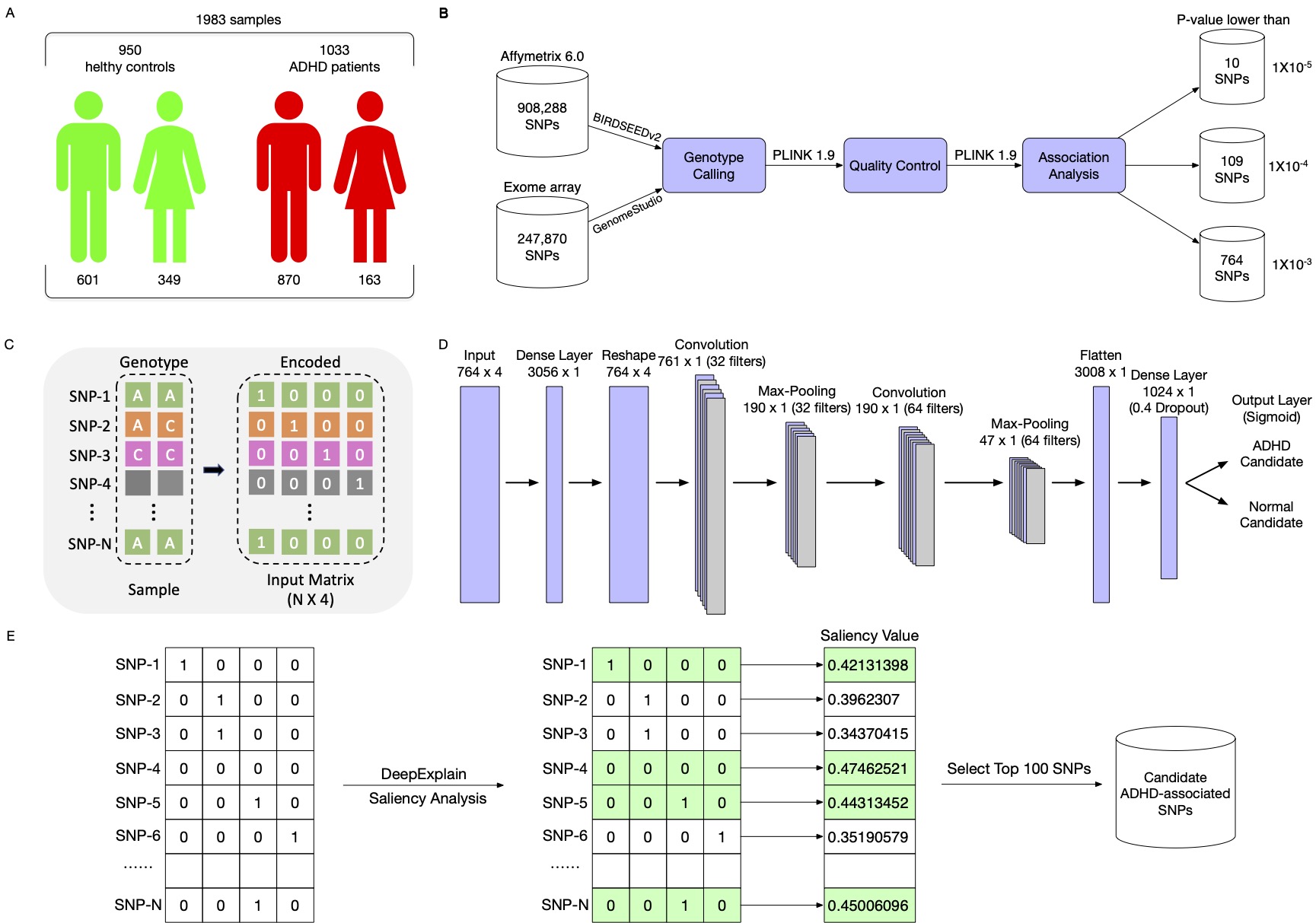
|
|
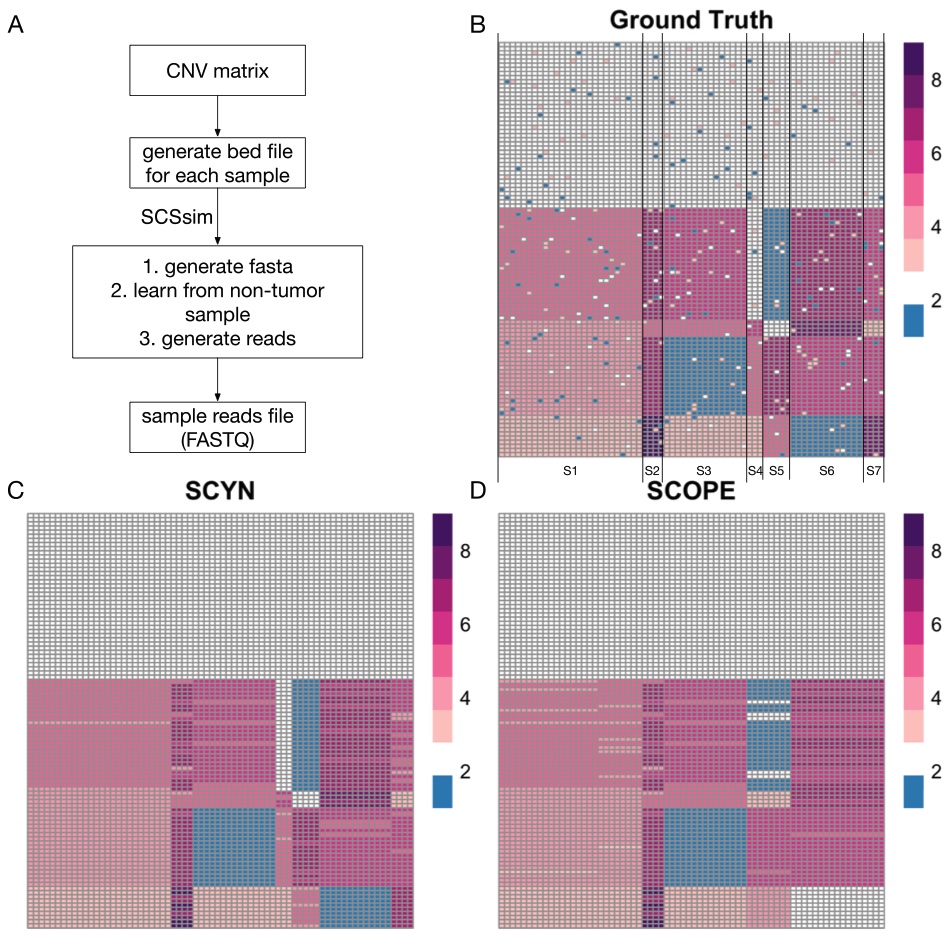
|
|
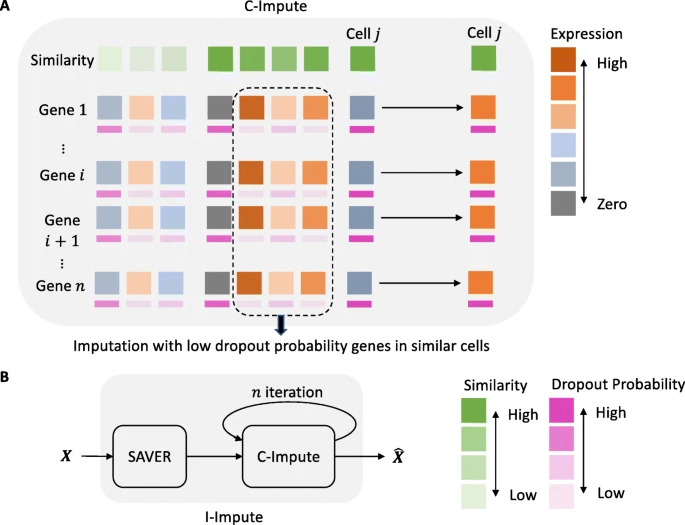
|
|
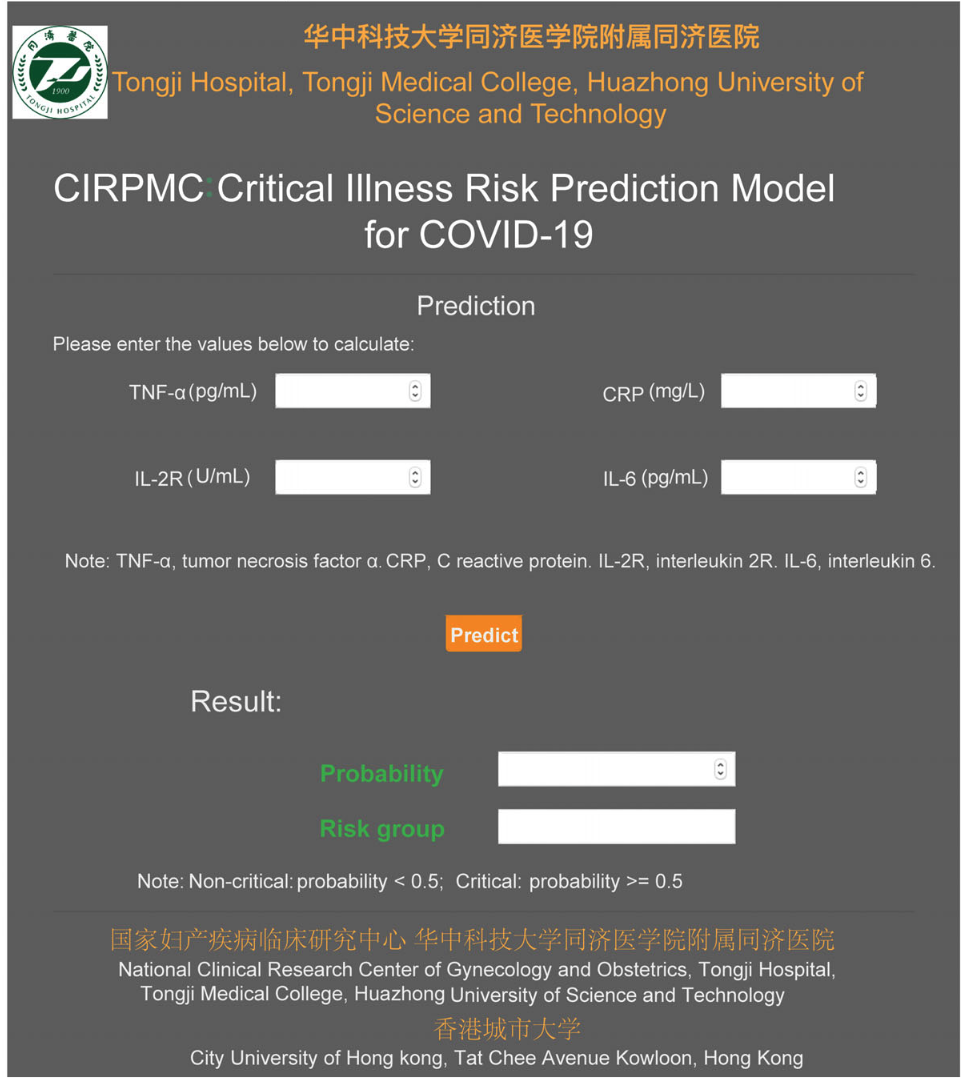
|
|
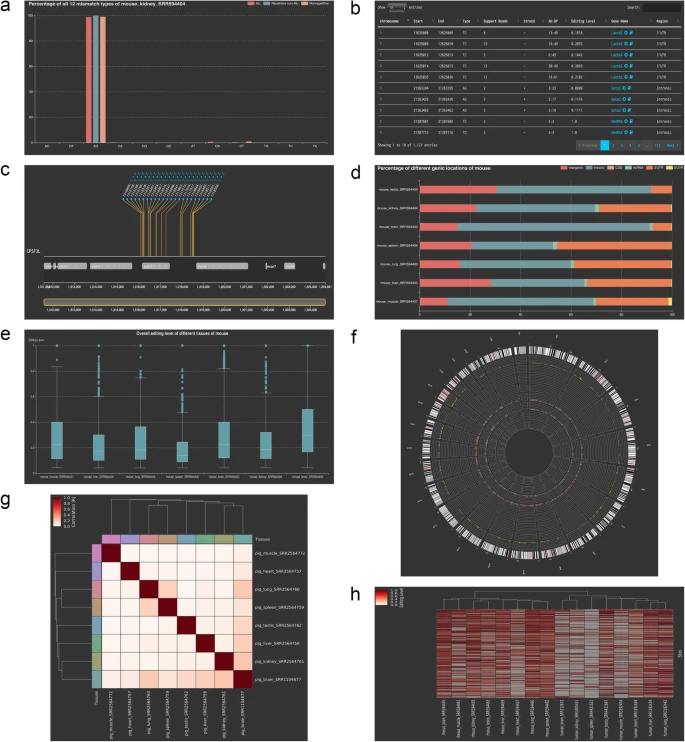
|
|
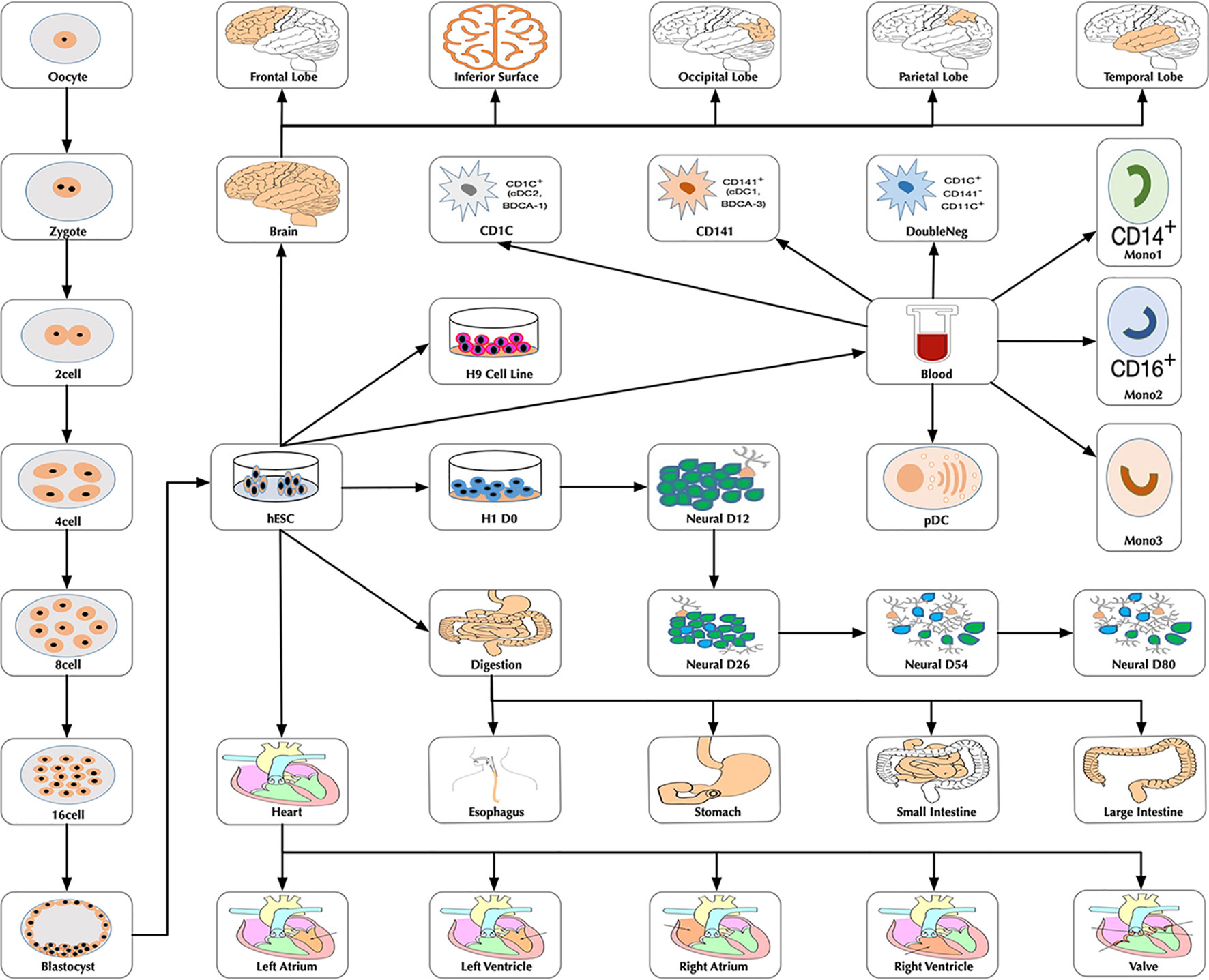
|
|
|
|
|
Last updated: Mar, 2025 by Xikang Feng
|